Example Views on the GRO Dataset Presented in Millstone
Demo Machine Front Page with project list.
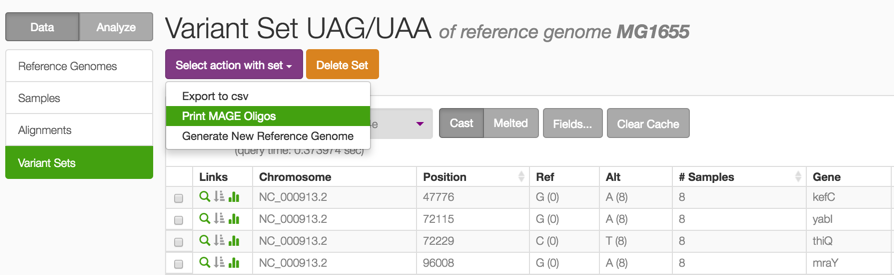
1. A list of all designed UAG-to-UAA mutations, in Variant Set view.
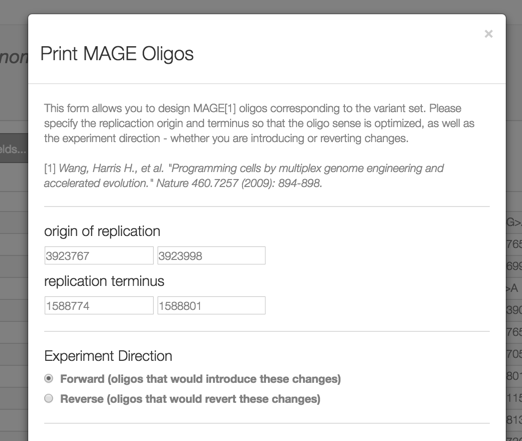
2. After clicking the link above, from the ‘Select action with set’ button, you can design MAGE oligonucleotides for each variant from this set, to either revert or create these mutations. This function is enabled in the demo, you can download a .csv with the oligos to synthesize.
3. List all variants which occur in the final strain, are not designed, and have a high effect. These are candidates for reversion to fix any phenotype deficits.
VARIANT_SET_UID=964875c3 & SAMPLE = recoli_misq_c31_321D & (INFO_EFF_IMPACT = HIGH | INFO_EFF_IMPACT = MODERATE) & INFO_EFF_EFFECT != None
4. List of tolC deletions in various strains, toggled during negative and positive selection during strain assembly.